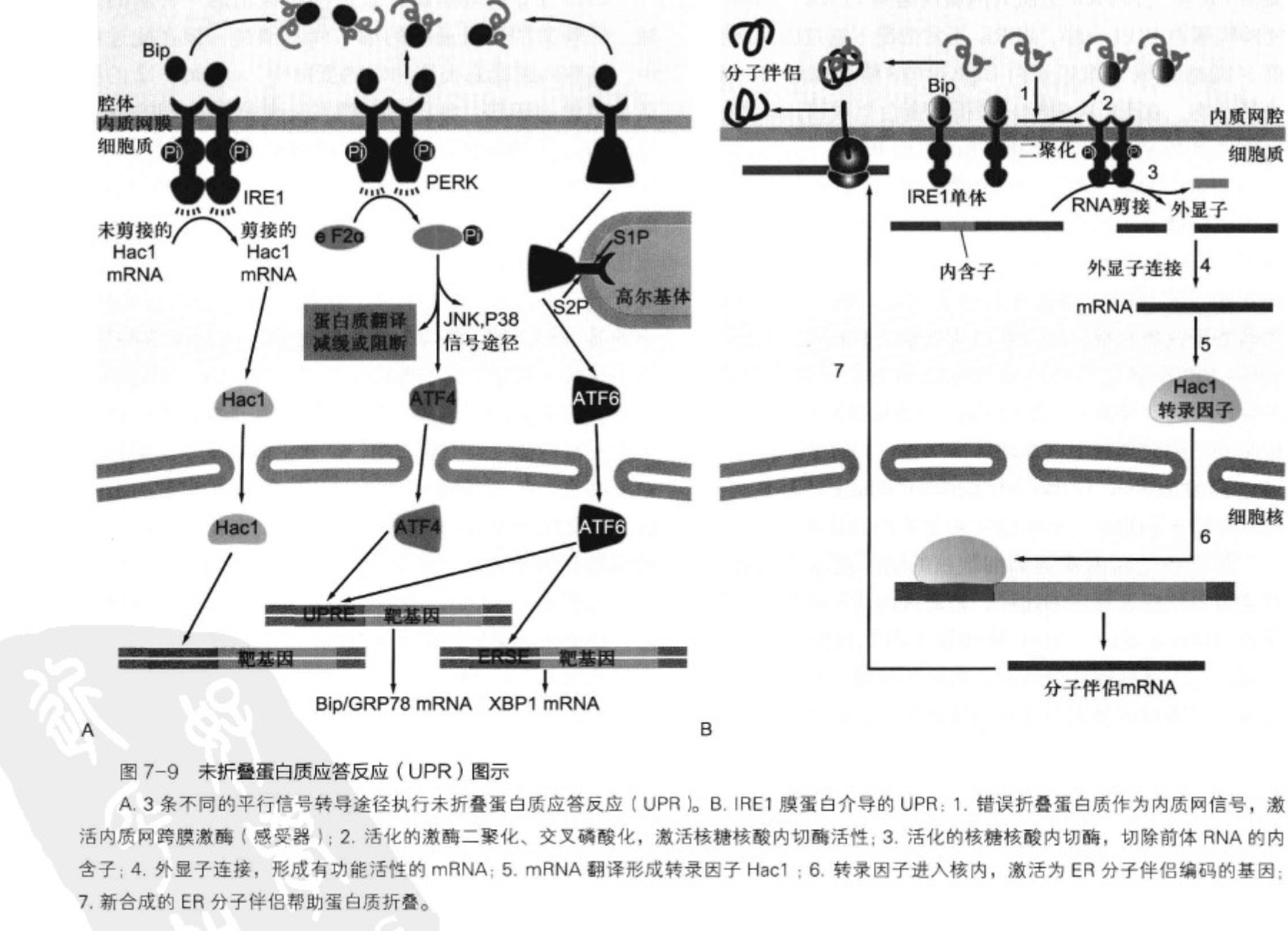
Unfolded Protein Response, UPR
- IRE1/HAC1
- PERK
- ATF6
eLife digest
Protein folding—the process by which a sequence of amino acids adopts the precise shape that is needed to perform a specific biological function—is one of the most important processes in all of biology. Any sequence of amino acids has the potential to fold into a large number of different shapes, and misfolded proteins can lead to toxicity and other problems. For example, all cells rely on signaling proteins in the membranes that enclose them to monitor their environment so that they can adapt to changing conditions and, in multicellular organisms, communicate with neighboring cells: without properly folded signaling proteins, chaos would ensue. Moreover, many diseases—including diabetes, cancer, viral infection and neurodegenerative disease—have been linked to protein folding processes. It is not surprising, therefore, that cells have evolved elaborate mechanisms to exert exquisite quality control over protein folding.
One of these mechanisms, called the unfolded protein response (UPR), operates in a compartment within the cell known as the endoplasmic reticulum (ER). The ER is a labyrinthine network of tubes and sacs within all eukaryotic cells, and most proteins destined for the cell surface or outside the cell adopt their properly folded shapes within this compartment. If the ER does not have enough capacity to fold all of the proteins that are delivered there, the UPR switches on to increase the protein folding capacity, to expand the surface area and volume of the compartment, and to degrade misfolded proteins. If the UPR cannot adequately adjust the folding capacity of the ER to meet the demands of the cell, the UPR triggers a program that kills the cell to prevent putting the whole organism at risk.
Researchers have identified the cellular components that monitor the protein folding conditions inside the ER. All eukaryotic cells, from unicellular yeasts to mammalian cells, contain a highly conserved protein-folding sensor called Ire1. In all species analyzed to date, Ire1 is known to activate the UPR through an messenger RNA (mRNA) splicing mechanism. This splicing event provides the switch that drives a gene expression program in which the production of ER components is increased to boost the protein folding capacity of the compartment.
Kimmig, Diaz et al. now report the first instance of an organism in which the UPR does not involve mRNA splicing or the initiation of a gene expression program. Rather, the yeast Schizosaccharomyces pombe utilizes Ire1 to an entirely different end. The authors find that the activation of Ire1 in S. pombe leads to the selective decay of a specific class of mRNAs that all encode proteins entering the ER. Thus, rather than increasing the protein folding capacity of the ER when faced with an increased protein folding load, S. pombe cells correct the imbalance by decreasing the load.
The authors also show that a lone mRNA—the mRNA that encodes the molecular chaperone BiP, which is one of the major protein-folding components in the ER—uniquely escapes this decay. Rather than being degraded, Ire1 truncates BiP mRNA and renders it more stable. By studying the UPR in a divergent organism, the authors shed new light on the evolution of a universally important process and illustrate how conserved machinery has been repurposed.
Kimmig P, Diaz M, Zheng J, Williams CC, Lang A, Aragón T, Li H, Walter P. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. Elife. 2012 Oct 15;1:e00048. doi: 10.7554/eLife.00048. PMID: 23066505; PMCID: PMC3470409.
Introduction
Homeostatic control mechanisms are essential to life, allowing cells to balance capacity and demand of numerous physiological processes. One such mechanism, the unfolded protein response (UPR), operates in all eukaryotic cells to adjust the protein folding capacity of the endoplasmic reticulum (ER) according to need. Environmental or physiological demands can lead to an imbalance between the protein folding load and the protein folding capacity in the ER lumen, resulting in an accumulation of unfolded or misfolded proteins, a condition termed ‘ER stress’ (Walter and Ron, 2011). When unmitigated, ER stress is toxic to cells and triggers cell death (Shore et al., 2011; Tabas and Ron, 2011; Hetz, 2012).
The UPR is a network of evolutionarily conserved signal transduction pathways that monitors the conditions in the ER lumen to induce a transcriptional response. In metazoan cells, three ER-resident transmembrane sensors, Ire1, PERK, ATF6 transmit information into the cytosol. Each sensor activates transcription factors that collaborate to drive expression of UPR target genes (Walter and Ron, 2011), including genes encoding ER-lumenal chaperones, such as BiP, an abundantly expressed Hsp70 family member.
Ire1 is a bifunctional transmembrane kinase/endoribonuclease that controls expression of the transcription factor XBP1 by a non-conventional splicing of its mRNA. Ire1 uses its ER-lumenal domain to detect unfolded proteins, and in response activates by homo-oligomerization, trans-autophosphorylation, and allosteric activation of its cytosolic nuclease modality (Korennykh et al., 2009; Gardner and Walter, 2011). Activated Ire1 cleaves the XBP1 mRNA at two discrete stem-loop structures, excising a short intron. The two severed exons are then ligated to produce spliced XBP1 mRNA, which because of a frame-shift induced by the splicing event, are translated to produce active XBP1 (Yoshida et al., 2001; Calfon et al., 2002).
Ire1 was first discovered in the budding yeast S. cerevisiae, where it constitutes the core machinery of the cells’ only UPR signaling pathway (Cox et al., 1993; Mori et al., 1993). S. cerevisiae Ire1 splices Hac1 mRNA, encoding the yeast ortholog of XBP1, by a mechanism that was later found conserved in all metazoan cells (Cox and Walter, 1996; Sidrauski et al., 1996). Ire1-mediated mRNA splicing therefore is considered to be the most evolutionary ancient branch of the UPR.
By first approximation, the three UPR branches collaborate to effect comprehensive transcriptional outputs, thereby enhancing the capacity of the ER according to need. PERK superimposes another layer of control by reducing the load of proteins entering the ER through translational control (Pavitt and Ron, 2012). Similarly, Ire1 is thought to play a dual role in UPR regulation. In particular, Hollien and Weissman (2006) first discovered in Drosophila cells that Ire1 induction not only results in splicing of XBP1 mRNA but also mediates enhanced mRNA breakdown. This output of Ire1 activation, termed ‘regulated Ire1-dependent decay’ (RIDD), is conserved in mammalian cells, but not in S. cerevisiae, where transcriptional control via Hac1 mRNA splicing remains the only known route of UPR signaling (Niwa et al., 2005; Han et al., 2009; Hollien et al., 2009). All identified RIDD target mRNAs are translated by membrane-bound ribosomes at the ER surface, where they are cleaved, most likely by Ire1 directly (Han et al., 2009; Hollien et al., 2009; Cross et al., 2012). Once nicked and no longer protected by their polyA tails and 5′ caps, mRNA fragments are quickly degraded by the RNA surveillance machinery (Hollien and Weissman, 2006; Garneau et al., 2007).
By contrast to the strictly conserved stem/loop structures found at Hac1/XBP1 mRNA splice sites (Gonzalez et al., 1999), RIDD target mRNAs do not contain easily recognizable features in common. Consequently, RIDD is thought to arise by a more promiscuous cleavage mode of Ire1. It is unclear whether RIDD is mediated by an alternate conformation of activated Ire1, or whether it arises in a specific Ire1 oligomerization state, as high-order oligomerization may serve to locally enhance low affinity interactions through avidity effects. RIDD cleavage reactions have been reconstituted in vitro with recombinantly expressed purified Ire1, lending support to the notion that Ire1’s endoribonuclease actvity, rather than another enzyme recruited to it, carries out the initial cleavage reaction (Lee et al., 2011; Cross et al., 2012).
Because of Ire1’s dual output, the physiological consequences of RIDD have been difficult to decipher. RIDD has been suggested to play cytoprotective roles, such as contributing to important feedback control on proinsulin expression in pancreatic beta-cells or protecting liver cells from acetaminophen toxicity by degrading the mRNAs encoding the cytochrome P450 variants responsible for the drug’s toxification (Lipson et al., 2008; Hur et al., 2012). RIDD has also been suggested to play cytotoxic roles as a major contributor driving cells into apoptosis after prolonged and unmitigated exposure to ER stress (Han et al., 2009).
Surprisingly, in the work presented here we found no evidence that Ire1 controls transcription in the UPR of Schizosaccharomyces pombe. Instead, in S. pombe Ire1 maintains ER homeostasis through two post-transcriptional mechanisms: it initiates RIDD of a large, select set of ER-targeted mRNAs and processes Bip1 mRNA in an unprecedented way, thereby stabilizing it. Our studies reveal an unforeseen evolutionary plasticity in maintaining ER homeostasis.
在human中,可能是RtcB负责把被Ire1切开的Hac1/Xbp1的外显子连接起来的。(酿酒酵母中是Trl1p和Tpt1p)
Endoplasmic reticulum overload response, EOR
SREBP
#### Apoptosis
衣霉素和DTT可以诱导UPR,后者的作用是组织内质网中氨基酸之间二硫键的形成。